Artificial Intelligence for Targeted Protein Degradation

What are targeted protein degraders
Targeted protein degraders are a family of novel, next-generation drugs. These drugs work by hijacking the different ways our bodies naturally break down proteins. These drugs solve an important set of issues that plague the most common drug type, namely small molecule drugs, such as their lack of specificity, their risk of inducing drug resistance and their need to target “druggable” proteins, which only represent 10-15% of all proteins in humans. Targeted protein degraders overcome all these issues through their remarkable mechanism.
The most well-known of targeted protein degraders are PROTACs. A PROTAC is a heterobifunctional molecule. Heterobifunctional molecules are two different protein-binding molecules connected together with a molecular linker in order to create one single chemical compound. The first protein a PROTAC binds to is an E3 Ubiquitin Ligase. This protein is a naturally occurring protein in all eukaryotes (including humans), and is responsible (among other things) for the natural degradation of proteins that are attached to it through intracellular proteolysis. The second protein that a PROTAC binds to then, is the one to be degraded, and is considered the protein of interest (POI). The POI is disease-specific and its presence causes either one or multiple abnormal biological processes. By naturally degrading these POIs, PROTACs aim to reduce or fully remove the abnormal biological processes linked to a disease.
The first PROTAC was reported in the literature in 2001 by Sakamoto et al. and the first small molecule PROTAC only in 2008 by Schneekloth et al. Since then, the field has evolved rapidly: The first two PROTAC entering clinical trials were introduced in 2019 for cancer treatment, and a dozen more were announced to do so in 2021. Furthermore, many different type of heterobifunctional targeted protein degraders have been discovered since, such as AUTACS that degrade proteins through Autophagy, LYTACS that degrade proteins through the Lysosome, or DUBTACS that degrade proteins through Deubiquitination. For the sake of completeness, two additional types of targeted protein degraders exist, namely Molecular Glues and Monovalent Degraders.
The benefits of Targeted Protein Degraders
This particular attention toward Targeted Protein Degraders is explained by their great potential as therapeutic agents :
-
Most of them proved to be surprisingly selective toward their POI;
-
They require low dosing since one Targeted Protein Degrader can tag many POI (in contrast with regular drugs that have to stay bound to their target in order to inhibit activity);
-
They can target proteins that, until now, have been considered as undruggable because they have no or very limited interaction sites;
-
They have the capacity to overcome resistance from small molecules because they both reduce the mutagenic effect they have on proteins and can also target mutated proteins.
-
While Targeted Protein Degraders are bigger than usual small molecules, many small molecule Targeted Protein Degraders are able to penetrate cells and have good biodistribution properties.
Despite all those promising properties of the Targeted Protein Degraders, many uncertainties and challenges still lie in the field :
-
Destroying a protein instead of inhibiting its activity might, in some cases, prove detrimental. Toxicity induced by targeted protein degraders is a very nascent field of study that will evolve greatly in the coming years.
-
Predicting whether a targeted protein degrader will penetrate a cell wall is still difficult, and little experimental data exists to make accurate predictions for new and unseen targeted protein degraders;
-
The number of ligands to the degrader protein (such as the E3 ligase ligand for PROTACs) is very limited and many of these ligands are not always efficient. The circumstances under which some cause ubiquitination and others don’t are not well understood.
-
The role of the linker is also not well understood and might play a critical role in the drug’s efficacy, and minor differences in the linkers may cause heterobifunctional targeted protein degraders to be either fully effective or not effective at all .
-
Finding target ligands is still an expensive, time-consuming problem, and which has traditionally been tackled through a “brute force search” in the form of high throughput screenings of the binding potential of hundreds of thousands of compounds.
-
The engineering of the full heterobifunctional targeted protein degraders by combining linkers, target ligands and ligase ligands involves many choices, and finding optimal configuration is still very much an art rather than a science.
In conclusion, there is great promise of using targeted protein degraders as efficient and target specific drugs but also still many challenges to overcome.
How Kantify uses AI to design targeted protein degraders
At Kantify, we use Artificial Intelligence in a number of ways to help in the discovery of safe and effective targeted protein degraders. These different algorithms are centralized in a single application called Sapian.
We use Sapian in three distinct ways: i. To find ligands to both the POI and the degrader, ii. To design the heterobifunctional targeted protein degraders in such a way that they maintain their binding capabilities, and iii. To predict the safety and effectiveness of these newly designed compounds.
How Sapian is used in PROTAC design
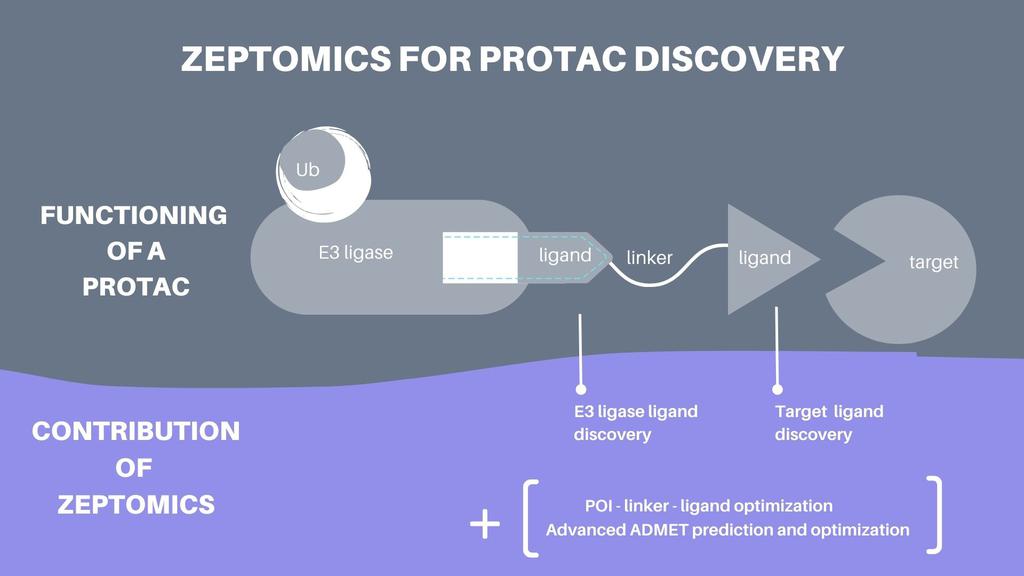
AI to identify ligands
As mentioned earlier, one needs to find ligands to both the POI and to the degrader in order to create heterobifunctional targeted protein degraders. Both of these tasks are challenging. Firstly, while some small molecule ligands to degraders are known from prior research, this list remains small, and the list of ligands that can be combined into effective targeted protein degraders is smaller still. Secondly, while most of the “druggable” POI have known ligands, these ligands aren’t always good candidates for targeted protein degraders, as they can be non-specific to the POI and/or have physical properties that make them unsuitable to be used in a heterobifunctional targeted protein degrader. POIs that are not druggable suffer from an even worse fate, as many don’t have any known ligands.
Traditionally, these problems are solved by running high throughput screens, which involves evaluating in vitro which of hundreds of thousands or even millions of compounds bind to the degrader or of the POI. Because of this “brute force” approach, this method is costly, slow and wasteful, taking up to 6 months and costing hundreds, if not millions of dollars to execute.
Kantify approaches this problem differently, by running an “in silico” screen, where a highly optimized Artificial Intelligence algorithm predicts whether a given compound is likely going to bind to a given protein, in a process that takes hours, rather than months, and that achieves close to in-vitro results. The handful of most promising candidates can then be tested in a lab at a fraction of the cost, time and effort.
AI to design drugs
In order to create a final targeted protein degrader that can be synthesized by a chemist, one has to select one of many POI ligands, one of a few degrader ligands, and one of many possible linkers. Sadly, most of these combinations experimentally prove to be ineffective, due to the fact they are mechanically hindered by each other, or that the placement causes them to lose their binding ability. Furthermore, and as mentioned previously, not all ligands and linkers combine well into safe and effective drugs.
Traditionally, these problems are solved by synthesizing many different possible combinations in order to find effective candidate drugs. Once more, this approach is costly, slow and wasteful, and can easily cost tens if not hundreds of thousands of dollars to execute.
Kantify approaches this problem computationally, by using its Artificial Intelligence algorithm to evaluate the binding ability of all possible combinations of POI ligands, degrader ligands and linkers to their respective targets. Thanks to this computational approach we quickly figure out which combinations are most optimal.
AI to evaluate safety and effectiveness
Finally, while a heterobifunctional targeted protein degrader can effectively bind to both of its targets, it can still remain an unsafe or non-effective drug for other reasons.
Traditionally, a variety of properties are evaluated either in vitro or in vivo models, and can include elements of absorption, distribution, metabolism, excretion and toxicological properties (ADMET) of the compounds. The evaluation of each ADMET endpoint is, once again, costly, slow and wasteful. And can cost tens to hundreds of thousands of dollars to execute. Kantify uses Artificial Intelligence to solve this problem, by predicting over 80 different ADMET properties in silico, reducing the cost, time and impact on animal life of an evaluation of a drug candidate, and allowing only candidates to be synthesized that have the best possible ADMET properties.
The path forward
We believe that the use of AI in drug discovery in general, and in the discovery of targeted protein degraders is still nascent and will continue to grow exponentially over the next few years. If you are interested in finding out more, please feel free to reach out to us at hello@kantify.com